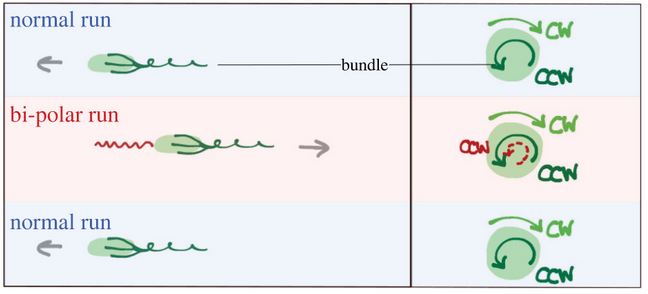
 In many situations, bacteria move in complex environments, as soils, oceans or the human gut-track, where carrier fluids show complex structures associated with non-Newtonian rheology. Many fundamental questions concerning the ability to navigate in such environments remain unsolved. Recently, it has been shown that the kinetics of bacterial motion in structured fluids as liquid crystals (LCs) is constrained by the orientational molecular order (or director field) and that novel spatio-temporal patterns arise. A question unaddressed so far is how bacteria change swimming direction in such an environment. In this work, we study the swimming mechanism of a single bacterium, Esherichia coli, constrained to move along the director field of a lyotropic chromonic liquid crystal confined to a planar cell. Here, the spontaneous ’run and tumble’ motion of the bacterium gets frustrated: the elasticity of the LC prevents flagella from unbundling. Interestingly, to change direction, bacteria execute a reversal motion along the director field, driven by the relocation of a single flagellum, a ’frustrated tumble’. We characterize this phenomenon in detail experimentally, exploiting exceptional spatial and temporal resolution of bacterial and flagellar dynamics, using a two colour Lagrangian tracking technique. We suggest a possible mechanism accounting for these observations.
In many situations, bacteria move in complex environments, as soils, oceans or the human gut-track, where carrier fluids show complex structures associated with non-Newtonian rheology. Many fundamental questions concerning the ability to navigate in such environments remain unsolved. Recently, it has been shown that the kinetics of bacterial motion in structured fluids as liquid crystals (LCs) is constrained by the orientational molecular order (or director field) and that novel spatio-temporal patterns arise. A question unaddressed so far is how bacteria change swimming direction in such an environment. In this work, we study the swimming mechanism of a single bacterium, Esherichia coli, constrained to move along the director field of a lyotropic chromonic liquid crystal confined to a planar cell. Here, the spontaneous ’run and tumble’ motion of the bacterium gets frustrated: the elasticity of the LC prevents flagella from unbundling. Interestingly, to change direction, bacteria execute a reversal motion along the director field, driven by the relocation of a single flagellum, a ’frustrated tumble’. We characterize this phenomenon in detail experimentally, exploiting exceptional spatial and temporal resolution of bacterial and flagellar dynamics, using a two colour Lagrangian tracking technique. We suggest a possible mechanism accounting for these observations.
(Abstract)
INTERFACE FOCUS
Volume12/ Issue6/ Article Number 20220039
Published OCT2022
By: Martyna Goral, Eric Clement, Thierry Darnige, Teresa Lopez-Leon, Anke Lindner

